Cloud execution on Google Colab#
Here, we will demo Neuroscout in cloud using the free resources available in Google Colab. You check it out as rendered here in a static version, run it interactively via the little rocket icon followed by clicking on/selecting Colab or directly via .
Specifically, this section will show you how you can run a sample pre-generated analysis with an already provided id or provide your own analysis id. To run your own analysis, copy your id into the field in the cell labelled (_1), set Neuroscout Analysis ID_ and then run all of the cells. The provided here id will run 10 subjects and 1 run from the Budapest dataset, and may take around 15 minutes. Larger analyses will take longer due to the limited free resources. To minimize downloads, we’ve uploaded the Budapest dataset to Google Drive, but feel free to run analyses from other datasets by downloading data using the get command.
In order to make this work here, we need to install a few things first. This includes Neurodebian (to easily get DataLad), DataLad (to get the data, here the Budapest dataset), and of course Neuroscout.
#@title 1) Run once to install Neuroscout-CLI & DataLad (~1 minute)
%%capture --no-display --no-stderr
from IPython.display import display
display("Installing NeuroDebian...")
## Set up DataLad
!wget -O- http://neuro.debian.net/lists/bionic.us-tn.full | sudo tee /etc/apt/sources.list.d/neurodebian.sources.list && sudo apt-key adv --recv-keys --keyserver hkps://keyserver.ubuntu.com 0xA5D32F012649A5A9 && sudo apt-get update
display("Installing DataLad...")
!sudo apt-get install datalad -y
!git config --global user.email "you@example.com" && git config --global user.name "Your Name"
display("Installing Neuroscout...")
%pip install neuroscout-cli
display("Done.")
'Installing NeuroDebian...'
'Installing DataLad...'
'Installing Neuroscout...'
'Done.'
Now we can set the analysis ID as outlined above.
#@title 2) Set your Neuroscout Analysis ID { run: "auto" }
analysis_id = 'iarv7' #@param {type:"string"}
import os
os.environ['ANALYSIS_ID'] = analysis_id
print(f"Analysis ID set to: {analysis_id}")
Analysis ID set to: iarv7
3) Mount Google Drive (optional)#
For this demo, we provide a copy of the Budapest dataset in Google Drive.
You can run this model, and any model using this dataset without downloading any fMRI data.
This is optional so feel free to skip this step, and the run command will download the required data for any analysis_id automatically.
First, use this link to add the
neuroscout-datasetsfolder to your Drive: https://drive.google.com/drive/folders/1wpw6wholT8ej-xhB-gsXo2tnahSvxCWs?usp=sharingMake a shorcut to the
neuroscout-datasetsfolder on your Google Drive (this will not use up any space on your Drive).Run this command to mount your Drive to this Notebook (You will have to approve permissions for this notebook to access your Drive).
from google.colab import drive
drive.mount('/content/drive')
Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount("/content/drive", force_remount=True).
4) Run your model#
Now, we can use neuroscout to run our analysis.
neuroscout uses FitLins to intepret your analysis’ BIDS Statistical Model, and construct a workflow to fit the model.
We can pass any valid FitLins arguments by passing them to neuroscout.
For example, --n-cpus=2 is passed to FitLins to set the maximum number of CPU cores (Colab only has 2 cores available).
Since we are providing a cached dataset, we will instruct neuroscout to skip data fetching (--no-get), and point neuroscout to the --download-dir where our cached data from Drive is.
!neuroscout run --no-get --download-dir=/content/drive/MyDrive/neuroscout-datasets --n-cpus=2 $ANALYSIS_ID /out
INFO:root:Analysis ID : iarv7
WARNING:cli:Captured warning (<class 'UserWarning'>): The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
IMPORTANT:cli:
Running FitLins version 0.10.1:
* Participant list: None.
WARNING:cli:Captured warning (<class 'UserWarning'>): [Node run; Contrast speech]:Contrast 'Type' is now 'Test'.
220429-20:13:39,423 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.loader" in "/tmp/tmpqslk4cp3/fitlins_wf/loader".
220429-20:13:39,427 nipype.workflow INFO:
[Node] Executing "loader" <fitlins.interfaces.bids.LoadBIDSModel>
/usr/local/lib/python3.7/dist-packages/bids/layout/validation.py:151: UserWarning: The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
warnings.warn("The PipelineDescription field was superseded "
/usr/local/lib/python3.7/dist-packages/bids/modeling/statsmodels.py:56: UserWarning: [Node run; Contrast speech]:Contrast 'Type' is now 'Test'.
warnings.warn(f"[Node {node['name']}; Contrast {contrast['name']}]:"
220429-20:13:44,800 nipype.workflow INFO:
[Node] Finished "loader", elapsed time 5.371779s.
220429-20:13:50,928 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.getter" in "/tmp/tmpqslk4cp3/fitlins_wf/getter".
220429-20:13:51,276 nipype.workflow INFO:
[Node] Executing "getter" <fitlins.interfaces.bids.BIDSSelect>
/usr/local/lib/python3.7/dist-packages/bids/layout/validation.py:151: UserWarning: The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
warnings.warn("The PipelineDescription field was superseded "
220429-20:13:52,844 nipype.workflow INFO:
[Node] Finished "getter", elapsed time 1.5667010000000001s.
220429-20:13:57,54 nipype.workflow INFO:
[Node] Setting-up "_design_matrix0" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix0".
220429-20:13:57,59 nipype.workflow INFO:
[Node] Executing "_design_matrix0" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:13:57,62 nipype.workflow INFO:
[Node] Setting-up "_design_matrix1" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix1".
220429-20:13:57,69 nipype.workflow INFO:
[Node] Executing "_design_matrix1" <fitlins.interfaces.nistats.DesignMatrix>
/usr/local/lib/python3.7/dist-packages/nilearn/datasets/__init__.py:96: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
"Numpy arrays.", FutureWarning)
/usr/local/lib/python3.7/dist-packages/nilearn/glm/__init__.py:56: FutureWarning: The nilearn.glm module is experimental. It may change in any future release of Nilearn.
'It may change in any future release of Nilearn.', FutureWarning)
220429-20:13:57,490 nipype.workflow INFO:
[Node] Finished "_design_matrix0", elapsed time 0.031583s.
/usr/local/lib/python3.7/dist-packages/nilearn/datasets/__init__.py:96: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
"Numpy arrays.", FutureWarning)
/usr/local/lib/python3.7/dist-packages/nilearn/glm/__init__.py:56: FutureWarning: The nilearn.glm module is experimental. It may change in any future release of Nilearn.
'It may change in any future release of Nilearn.', FutureWarning)
220429-20:13:57,929 nipype.workflow INFO:
[Node] Finished "_design_matrix1", elapsed time 0.063748s.
220429-20:13:59,52 nipype.workflow INFO:
[Node] Setting-up "_design_matrix2" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix2".
220429-20:13:59,58 nipype.workflow INFO:
[Node] Executing "_design_matrix2" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:13:59,58 nipype.workflow INFO:
[Node] Setting-up "_design_matrix3" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix3".
220429-20:13:59,67 nipype.workflow INFO:
[Node] Executing "_design_matrix3" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:13:59,88 nipype.workflow INFO:
[Node] Finished "_design_matrix2", elapsed time 0.028413s.
220429-20:13:59,92 nipype.workflow INFO:
[Node] Finished "_design_matrix3", elapsed time 0.024257s.
220429-20:14:01,56 nipype.workflow INFO:
[Node] Setting-up "_design_matrix4" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix4".
220429-20:14:01,57 nipype.workflow INFO:
[Node] Setting-up "_design_matrix5" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix5".
220429-20:14:01,62 nipype.workflow INFO:
[Node] Executing "_design_matrix5" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:01,65 nipype.workflow INFO:
[Node] Executing "_design_matrix4" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:01,85 nipype.workflow INFO:
[Node] Finished "_design_matrix5", elapsed time 0.021781s.
220429-20:14:01,93 nipype.workflow INFO:
[Node] Finished "_design_matrix4", elapsed time 0.027267s.
220429-20:14:03,54 nipype.workflow INFO:
[Node] Setting-up "_design_matrix7" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix7".
220429-20:14:03,56 nipype.workflow INFO:
[Node] Setting-up "_design_matrix6" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix6".
220429-20:14:03,59 nipype.workflow INFO:
[Node] Executing "_design_matrix7" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:03,63 nipype.workflow INFO:
[Node] Executing "_design_matrix6" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:03,86 nipype.workflow INFO:
[Node] Finished "_design_matrix7", elapsed time 0.025186s.
220429-20:14:03,88 nipype.workflow INFO:
[Node] Finished "_design_matrix6", elapsed time 0.021832s.
220429-20:14:05,55 nipype.workflow INFO:
[Node] Setting-up "_design_matrix8" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix8".
220429-20:14:05,60 nipype.workflow INFO:
[Node] Executing "_design_matrix8" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:05,61 nipype.workflow INFO:
[Node] Setting-up "_design_matrix9" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix9".
220429-20:14:05,75 nipype.workflow INFO:
[Node] Executing "_design_matrix9" <fitlins.interfaces.nistats.DesignMatrix>
220429-20:14:05,83 nipype.workflow INFO:
[Node] Finished "_design_matrix8", elapsed time 0.021489s.
220429-20:14:05,95 nipype.workflow INFO:
[Node] Finished "_design_matrix9", elapsed time 0.018316s.
220429-20:14:07,105 nipype.workflow INFO:
[Node] Setting-up "_design_matrix0" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix0".
220429-20:14:07,108 nipype.workflow INFO:
[Node] Cached "_design_matrix0" - collecting precomputed outputs
220429-20:14:07,108 nipype.workflow INFO:
[Node] "_design_matrix0" found cached.
220429-20:14:07,110 nipype.workflow INFO:
[Node] Setting-up "_design_matrix1" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix1".
220429-20:14:07,113 nipype.workflow INFO:
[Node] Cached "_design_matrix1" - collecting precomputed outputs
220429-20:14:07,113 nipype.workflow INFO:
[Node] "_design_matrix1" found cached.
220429-20:14:07,115 nipype.workflow INFO:
[Node] Setting-up "_design_matrix2" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix2".
220429-20:14:07,117 nipype.workflow INFO:
[Node] Cached "_design_matrix2" - collecting precomputed outputs
220429-20:14:07,118 nipype.workflow INFO:
[Node] "_design_matrix2" found cached.
220429-20:14:07,119 nipype.workflow INFO:
[Node] Setting-up "_design_matrix3" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix3".
220429-20:14:07,123 nipype.workflow INFO:
[Node] Cached "_design_matrix3" - collecting precomputed outputs
220429-20:14:07,123 nipype.workflow INFO:
[Node] "_design_matrix3" found cached.
220429-20:14:07,124 nipype.workflow INFO:
[Node] Setting-up "_design_matrix4" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix4".
220429-20:14:07,127 nipype.workflow INFO:
[Node] Cached "_design_matrix4" - collecting precomputed outputs
220429-20:14:07,127 nipype.workflow INFO:
[Node] "_design_matrix4" found cached.
220429-20:14:07,128 nipype.workflow INFO:
[Node] Setting-up "_design_matrix5" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix5".
220429-20:14:07,131 nipype.workflow INFO:
[Node] Cached "_design_matrix5" - collecting precomputed outputs
220429-20:14:07,131 nipype.workflow INFO:
[Node] "_design_matrix5" found cached.
220429-20:14:07,133 nipype.workflow INFO:
[Node] Setting-up "_design_matrix6" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix6".
220429-20:14:07,135 nipype.workflow INFO:
[Node] Cached "_design_matrix6" - collecting precomputed outputs
220429-20:14:07,135 nipype.workflow INFO:
[Node] "_design_matrix6" found cached.
220429-20:14:07,137 nipype.workflow INFO:
[Node] Setting-up "_design_matrix7" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix7".
220429-20:14:07,140 nipype.workflow INFO:
[Node] Cached "_design_matrix7" - collecting precomputed outputs
220429-20:14:07,140 nipype.workflow INFO:
[Node] "_design_matrix7" found cached.
220429-20:14:07,142 nipype.workflow INFO:
[Node] Setting-up "_design_matrix8" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix8".
220429-20:14:07,145 nipype.workflow INFO:
[Node] Cached "_design_matrix8" - collecting precomputed outputs
220429-20:14:07,145 nipype.workflow INFO:
[Node] "_design_matrix8" found cached.
220429-20:14:07,146 nipype.workflow INFO:
[Node] Setting-up "_design_matrix9" in "/tmp/tmpqslk4cp3/fitlins_wf/design_matrix/mapflow/_design_matrix9".
220429-20:14:07,149 nipype.workflow INFO:
[Node] Cached "_design_matrix9" - collecting precomputed outputs
220429-20:14:07,149 nipype.workflow INFO:
[Node] "_design_matrix9" found cached.
220429-20:14:11,260 nipype.workflow INFO:
[Node] Setting-up "_plot_design0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design0".
220429-20:14:11,262 nipype.workflow INFO:
[Node] Executing "_plot_design0" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:11,264 nipype.workflow INFO:
[Node] Setting-up "_plot_design1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design1".
220429-20:14:11,266 nipype.workflow INFO:
[Node] Executing "_plot_design1" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:11,699 nipype.workflow INFO:
[Node] Finished "_plot_design1", elapsed time 0.431299s.
220429-20:14:11,699 nipype.workflow INFO:
[Node] Finished "_plot_design0", elapsed time 0.435562s.
220429-20:14:13,63 nipype.workflow INFO:
[Node] Setting-up "_plot_design2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design2".
220429-20:14:13,65 nipype.workflow INFO:
[Node] Setting-up "_plot_design3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design3".
220429-20:14:13,66 nipype.workflow INFO:
[Node] Executing "_plot_design2" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:13,67 nipype.workflow INFO:
[Node] Executing "_plot_design3" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:13,433 nipype.workflow INFO:
[Node] Finished "_plot_design2", elapsed time 0.366273s.
220429-20:14:13,463 nipype.workflow INFO:
[Node] Finished "_plot_design3", elapsed time 0.394361s.
220429-20:14:15,65 nipype.workflow INFO:
[Node] Setting-up "_plot_design4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design4".
220429-20:14:15,67 nipype.workflow INFO:
[Node] Setting-up "_plot_design5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design5".
220429-20:14:15,68 nipype.workflow INFO:
[Node] Executing "_plot_design4" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:15,70 nipype.workflow INFO:
[Node] Executing "_plot_design5" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:15,314 nipype.workflow INFO:
[Node] Finished "_plot_design5", elapsed time 0.24323s.
220429-20:14:15,540 nipype.workflow INFO:
[Node] Finished "_plot_design4", elapsed time 0.47065s.
220429-20:14:17,69 nipype.workflow INFO:
[Node] Setting-up "_plot_design6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design6".
220429-20:14:17,70 nipype.workflow INFO:
[Node] Setting-up "_plot_design7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design7".
220429-20:14:17,71 nipype.workflow INFO:
[Node] Executing "_plot_design6" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:17,73 nipype.workflow INFO:
[Node] Executing "_plot_design7" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:17,392 nipype.workflow INFO:
[Node] Finished "_plot_design6", elapsed time 0.320072s.
220429-20:14:17,480 nipype.workflow INFO:
[Node] Finished "_plot_design7", elapsed time 0.40633s.
220429-20:14:19,73 nipype.workflow INFO:
[Node] Setting-up "_plot_design8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design8".
220429-20:14:19,74 nipype.workflow INFO:
[Node] Setting-up "_plot_design9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design9".
220429-20:14:19,76 nipype.workflow INFO:
[Node] Executing "_plot_design8" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:19,77 nipype.workflow INFO:
[Node] Executing "_plot_design9" <fitlins.interfaces.visualizations.DesignPlot>
220429-20:14:19,372 nipype.workflow INFO:
[Node] Finished "_plot_design9", elapsed time 0.294205s.
220429-20:14:19,458 nipype.workflow INFO:
[Node] Finished "_plot_design8", elapsed time 0.380556s.
220429-20:14:21,82 nipype.workflow INFO:
[Node] Setting-up "_plot_design0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design0".
220429-20:14:21,87 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix0".
220429-20:14:21,88 nipype.workflow INFO:
[Node] Cached "_plot_design0" - collecting precomputed outputs
220429-20:14:21,88 nipype.workflow INFO:
[Node] "_plot_design0" found cached.
220429-20:14:21,90 nipype.workflow INFO:
[Node] Setting-up "_plot_design1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design1".
220429-20:14:21,91 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix0" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:21,91 nipype.workflow INFO:
[Node] Cached "_plot_design1" - collecting precomputed outputs
220429-20:14:21,91 nipype.workflow INFO:
[Node] "_plot_design1" found cached.
220429-20:14:21,92 nipype.workflow INFO:
[Node] Setting-up "_plot_design2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design2".
220429-20:14:21,93 nipype.workflow INFO:
[Node] Cached "_plot_design2" - collecting precomputed outputs
220429-20:14:21,94 nipype.workflow INFO:
[Node] "_plot_design2" found cached.
220429-20:14:21,95 nipype.workflow INFO:
[Node] Setting-up "_plot_design3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design3".
220429-20:14:21,96 nipype.workflow INFO:
[Node] Cached "_plot_design3" - collecting precomputed outputs
220429-20:14:21,96 nipype.workflow INFO:
[Node] "_plot_design3" found cached.
220429-20:14:21,97 nipype.workflow INFO:
[Node] Setting-up "_plot_design4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design4".
220429-20:14:21,98 nipype.workflow INFO:
[Node] Cached "_plot_design4" - collecting precomputed outputs
220429-20:14:21,99 nipype.workflow INFO:
[Node] "_plot_design4" found cached.
220429-20:14:21,100 nipype.workflow INFO:
[Node] Setting-up "_plot_design5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design5".
220429-20:14:21,101 nipype.workflow INFO:
[Node] Cached "_plot_design5" - collecting precomputed outputs
220429-20:14:21,101 nipype.workflow INFO:
[Node] "_plot_design5" found cached.
220429-20:14:21,102 nipype.workflow INFO:
[Node] Setting-up "_plot_design6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design6".
220429-20:14:21,103 nipype.workflow INFO:
[Node] Cached "_plot_design6" - collecting precomputed outputs
220429-20:14:21,104 nipype.workflow INFO:
[Node] "_plot_design6" found cached.
220429-20:14:21,105 nipype.workflow INFO:
[Node] Setting-up "_plot_design7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design7".
220429-20:14:21,106 nipype.workflow INFO:
[Node] Cached "_plot_design7" - collecting precomputed outputs
220429-20:14:21,106 nipype.workflow INFO:
[Node] "_plot_design7" found cached.
220429-20:14:21,107 nipype.workflow INFO:
[Node] Setting-up "_plot_design8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design8".
220429-20:14:21,109 nipype.workflow INFO:
[Node] Cached "_plot_design8" - collecting precomputed outputs
220429-20:14:21,109 nipype.workflow INFO:
[Node] "_plot_design8" found cached.
220429-20:14:21,110 nipype.workflow INFO:
[Node] Setting-up "_plot_design9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_design/mapflow/_plot_design9".
220429-20:14:21,111 nipype.workflow INFO:
[Node] Cached "_plot_design9" - collecting precomputed outputs
220429-20:14:21,111 nipype.workflow INFO:
[Node] "_plot_design9" found cached.
220429-20:14:21,282 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix0", elapsed time 0.190126s.
220429-20:14:23,93 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix1".
220429-20:14:23,94 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix2".
220429-20:14:23,96 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix1" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:23,97 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix2" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:23,519 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix1", elapsed time 0.418097s.
220429-20:14:23,599 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix2", elapsed time 0.501275s.
220429-20:14:25,75 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix3".
220429-20:14:25,77 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix4".
220429-20:14:25,79 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix3" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:25,80 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix4" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:25,488 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix4", elapsed time 0.404563s.
220429-20:14:25,492 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix3", elapsed time 0.41173s.
220429-20:14:27,79 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix5".
220429-20:14:27,81 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix6".
220429-20:14:27,83 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix5" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:27,84 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix6" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:27,469 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix5", elapsed time 0.378918s.
220429-20:14:27,481 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix6", elapsed time 0.395185s.
220429-20:14:29,76 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix7".
220429-20:14:29,78 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix8".
220429-20:14:29,80 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix7" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:29,81 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix8" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:29,477 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix7", elapsed time 0.396085s.
220429-20:14:29,478 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix8", elapsed time 0.396272s.
220429-20:14:31,82 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix9".
220429-20:14:31,83 nipype.workflow INFO:
[Node] Setting-up "_plot_corr0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr0".
220429-20:14:31,85 nipype.workflow INFO:
[Node] Executing "_plot_run_contrast_matrix9" <fitlins.interfaces.visualizations.ContrastMatrixPlot>
220429-20:14:31,86 nipype.workflow INFO:
[Node] Executing "_plot_corr0" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:31,484 nipype.workflow INFO:
[Node] Finished "_plot_run_contrast_matrix9", elapsed time 0.397116s.
220429-20:14:31,529 nipype.workflow INFO:
[Node] Finished "_plot_corr0", elapsed time 0.441665s.
220429-20:14:33,88 nipype.workflow INFO:
[Node] Setting-up "_plot_corr1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr1".
220429-20:14:33,91 nipype.workflow INFO:
[Node] Executing "_plot_corr1" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:33,96 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix0".
220429-20:14:33,98 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix0" - collecting precomputed outputs
220429-20:14:33,98 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix0" found cached.
220429-20:14:33,99 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix1".
220429-20:14:33,100 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix1" - collecting precomputed outputs
220429-20:14:33,101 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix1" found cached.
220429-20:14:33,102 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix2".
220429-20:14:33,103 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix2" - collecting precomputed outputs
220429-20:14:33,103 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix2" found cached.
220429-20:14:33,104 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix3".
220429-20:14:33,106 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix3" - collecting precomputed outputs
220429-20:14:33,106 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix3" found cached.
220429-20:14:33,107 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix4".
220429-20:14:33,108 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix4" - collecting precomputed outputs
220429-20:14:33,109 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix4" found cached.
220429-20:14:33,110 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix5".
220429-20:14:33,111 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix5" - collecting precomputed outputs
220429-20:14:33,111 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix5" found cached.
220429-20:14:33,112 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix6".
220429-20:14:33,114 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix6" - collecting precomputed outputs
220429-20:14:33,114 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix6" found cached.
220429-20:14:33,116 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix7".
220429-20:14:33,118 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix7" - collecting precomputed outputs
220429-20:14:33,118 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix7" found cached.
220429-20:14:33,119 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix8".
220429-20:14:33,121 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix8" - collecting precomputed outputs
220429-20:14:33,121 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix8" found cached.
220429-20:14:33,122 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrast_matrix9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrast_matrix/mapflow/_plot_run_contrast_matrix9".
220429-20:14:33,124 nipype.workflow INFO:
[Node] Cached "_plot_run_contrast_matrix9" - collecting precomputed outputs
220429-20:14:33,124 nipype.workflow INFO:
[Node] "_plot_run_contrast_matrix9" found cached.
220429-20:14:33,355 nipype.workflow INFO:
[Node] Finished "_plot_corr1", elapsed time 0.262654s.
220429-20:14:35,105 nipype.workflow INFO:
[Node] Setting-up "_plot_corr2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr2".
220429-20:14:35,107 nipype.workflow INFO:
[Node] Setting-up "_plot_corr3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr3".
220429-20:14:35,108 nipype.workflow INFO:
[Node] Executing "_plot_corr2" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:35,111 nipype.workflow INFO:
[Node] Executing "_plot_corr3" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:35,623 nipype.workflow INFO:
[Node] Finished "_plot_corr3", elapsed time 0.511568s.
220429-20:14:35,621 nipype.workflow INFO:
[Node] Finished "_plot_corr2", elapsed time 0.511764s.
220429-20:14:37,91 nipype.workflow INFO:
[Node] Setting-up "_plot_corr4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr4".
220429-20:14:37,93 nipype.workflow INFO:
[Node] Setting-up "_plot_corr5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr5".
220429-20:14:37,94 nipype.workflow INFO:
[Node] Executing "_plot_corr4" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:37,96 nipype.workflow INFO:
[Node] Executing "_plot_corr5" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:37,610 nipype.workflow INFO:
[Node] Finished "_plot_corr4", elapsed time 0.513795s.
220429-20:14:37,618 nipype.workflow INFO:
[Node] Finished "_plot_corr5", elapsed time 0.520913s.
220429-20:14:39,102 nipype.workflow INFO:
[Node] Setting-up "_plot_corr6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr6".
220429-20:14:39,103 nipype.workflow INFO:
[Node] Setting-up "_plot_corr7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr7".
220429-20:14:39,105 nipype.workflow INFO:
[Node] Executing "_plot_corr6" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:39,106 nipype.workflow INFO:
[Node] Executing "_plot_corr7" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:39,650 nipype.workflow INFO:
[Node] Finished "_plot_corr7", elapsed time 0.538538s.
220429-20:14:39,656 nipype.workflow INFO:
[Node] Finished "_plot_corr6", elapsed time 0.548744s.
220429-20:14:41,97 nipype.workflow INFO:
[Node] Setting-up "_plot_corr8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr8".
220429-20:14:41,98 nipype.workflow INFO:
[Node] Setting-up "_plot_corr9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr9".
220429-20:14:41,100 nipype.workflow INFO:
[Node] Executing "_plot_corr8" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:41,101 nipype.workflow INFO:
[Node] Executing "_plot_corr9" <fitlins.interfaces.visualizations.DesignCorrelationPlot>
220429-20:14:41,602 nipype.workflow INFO:
[Node] Finished "_plot_corr9", elapsed time 0.498786s.
220429-20:14:41,639 nipype.workflow INFO:
[Node] Finished "_plot_corr8", elapsed time 0.537771s.
220429-20:14:43,100 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv0" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv0".
220429-20:14:43,103 nipype.workflow INFO:
[Node] Executing "_deindex_tsv0" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:43,108 nipype.workflow INFO:
[Node] Setting-up "_plot_corr0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr0".
220429-20:14:43,109 nipype.workflow INFO:
[Node] Cached "_plot_corr0" - collecting precomputed outputs
220429-20:14:43,109 nipype.workflow INFO:
[Node] "_plot_corr0" found cached.
220429-20:14:43,111 nipype.workflow INFO:
[Node] Setting-up "_plot_corr1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr1".
220429-20:14:43,112 nipype.workflow INFO:
[Node] Cached "_plot_corr1" - collecting precomputed outputs
220429-20:14:43,113 nipype.workflow INFO:
[Node] "_plot_corr1" found cached.
220429-20:14:43,114 nipype.workflow INFO:
[Node] Setting-up "_plot_corr2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr2".
220429-20:14:43,115 nipype.workflow INFO:
[Node] Cached "_plot_corr2" - collecting precomputed outputs
220429-20:14:43,115 nipype.workflow INFO:
[Node] "_plot_corr2" found cached.
220429-20:14:43,116 nipype.workflow INFO:
[Node] Finished "_deindex_tsv0", elapsed time 0.010888s.
220429-20:14:43,116 nipype.workflow INFO:
[Node] Setting-up "_plot_corr3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr3".
220429-20:14:43,118 nipype.workflow INFO:
[Node] Cached "_plot_corr3" - collecting precomputed outputs
220429-20:14:43,118 nipype.workflow INFO:
[Node] "_plot_corr3" found cached.
220429-20:14:43,119 nipype.workflow INFO:
[Node] Setting-up "_plot_corr4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr4".
220429-20:14:43,120 nipype.workflow INFO:
[Node] Cached "_plot_corr4" - collecting precomputed outputs
220429-20:14:43,120 nipype.workflow INFO:
[Node] "_plot_corr4" found cached.
220429-20:14:43,121 nipype.workflow INFO:
[Node] Setting-up "_plot_corr5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr5".
220429-20:14:43,121 nipype.workflow INFO:
[Node] Cached "_plot_corr5" - collecting precomputed outputs
220429-20:14:43,122 nipype.workflow INFO:
[Node] "_plot_corr5" found cached.
220429-20:14:43,122 nipype.workflow INFO:
[Node] Setting-up "_plot_corr6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr6".
220429-20:14:43,123 nipype.workflow INFO:
[Node] Cached "_plot_corr6" - collecting precomputed outputs
220429-20:14:43,123 nipype.workflow INFO:
[Node] "_plot_corr6" found cached.
220429-20:14:43,124 nipype.workflow INFO:
[Node] Setting-up "_plot_corr7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr7".
220429-20:14:43,125 nipype.workflow INFO:
[Node] Cached "_plot_corr7" - collecting precomputed outputs
220429-20:14:43,125 nipype.workflow INFO:
[Node] "_plot_corr7" found cached.
220429-20:14:43,125 nipype.workflow INFO:
[Node] Setting-up "_plot_corr8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr8".
220429-20:14:43,126 nipype.workflow INFO:
[Node] Cached "_plot_corr8" - collecting precomputed outputs
220429-20:14:43,126 nipype.workflow INFO:
[Node] "_plot_corr8" found cached.
220429-20:14:43,127 nipype.workflow INFO:
[Node] Setting-up "_plot_corr9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_corr/mapflow/_plot_corr9".
220429-20:14:43,128 nipype.workflow INFO:
[Node] Cached "_plot_corr9" - collecting precomputed outputs
220429-20:14:43,128 nipype.workflow INFO:
[Node] "_plot_corr9" found cached.
220429-20:14:45,118 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv1" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv1".
220429-20:14:45,121 nipype.workflow INFO:
[Node] Executing "_deindex_tsv1" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:45,127 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv2" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv2".
220429-20:14:45,129 nipype.workflow INFO:
[Node] Executing "_deindex_tsv2" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:45,134 nipype.workflow INFO:
[Node] Finished "_deindex_tsv1", elapsed time 0.011662s.
220429-20:14:45,140 nipype.workflow INFO:
[Node] Finished "_deindex_tsv2", elapsed time 0.009466s.
220429-20:14:47,97 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv3" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv3".
220429-20:14:47,97 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv4" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv4".
220429-20:14:47,100 nipype.workflow INFO:
[Node] Executing "_deindex_tsv4" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:47,101 nipype.workflow INFO:
[Node] Executing "_deindex_tsv3" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:47,112 nipype.workflow INFO:
[Node] Finished "_deindex_tsv3", elapsed time 0.009972s.
220429-20:14:47,112 nipype.workflow INFO:
[Node] Finished "_deindex_tsv4", elapsed time 0.010615s.
220429-20:14:49,99 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv5" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv5".
220429-20:14:49,101 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv6" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv6".
220429-20:14:49,102 nipype.workflow INFO:
[Node] Executing "_deindex_tsv5" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:49,104 nipype.workflow INFO:
[Node] Executing "_deindex_tsv6" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:49,113 nipype.workflow INFO:
[Node] Finished "_deindex_tsv5", elapsed time 0.010424s.
220429-20:14:49,115 nipype.workflow INFO:
[Node] Finished "_deindex_tsv6", elapsed time 0.010192s.
220429-20:14:51,100 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv7" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv7".
220429-20:14:51,102 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv8" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv8".
220429-20:14:51,104 nipype.workflow INFO:
[Node] Executing "_deindex_tsv7" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:51,104 nipype.workflow INFO:
[Node] Executing "_deindex_tsv8" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:51,116 nipype.workflow INFO:
[Node] Finished "_deindex_tsv7", elapsed time 0.010488s.
220429-20:14:51,116 nipype.workflow INFO:
[Node] Finished "_deindex_tsv8", elapsed time 0.010358s.
220429-20:14:53,100 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv9" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv9".
220429-20:14:53,104 nipype.workflow INFO:
[Node] Executing "_deindex_tsv9" <nipype.interfaces.utility.wrappers.Function>
220429-20:14:53,105 nipype.workflow INFO:
[Node] Setting-up "_l1_model0" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model0".
220429-20:14:53,124 nipype.workflow INFO:
[Node] Finished "_deindex_tsv9", elapsed time 0.017059s.
220429-20:14:53,143 nipype.workflow INFO:
[Node] Executing "_l1_model0" <fitlins.interfaces.nistats.FirstLevelModel>
220429-20:14:55,168 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv0" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv0".
220429-20:14:55,170 nipype.workflow INFO:
[Node] Cached "_deindex_tsv0" - collecting precomputed outputs
220429-20:14:55,170 nipype.workflow INFO:
[Node] "_deindex_tsv0" found cached.
220429-20:14:55,172 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv1" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv1".
220429-20:14:55,173 nipype.workflow INFO:
[Node] Cached "_deindex_tsv1" - collecting precomputed outputs
220429-20:14:55,173 nipype.workflow INFO:
[Node] "_deindex_tsv1" found cached.
220429-20:14:55,175 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv2" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv2".
220429-20:14:55,176 nipype.workflow INFO:
[Node] Cached "_deindex_tsv2" - collecting precomputed outputs
220429-20:14:55,177 nipype.workflow INFO:
[Node] "_deindex_tsv2" found cached.
220429-20:14:55,178 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv3" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv3".
220429-20:14:55,180 nipype.workflow INFO:
[Node] Cached "_deindex_tsv3" - collecting precomputed outputs
220429-20:14:55,180 nipype.workflow INFO:
[Node] "_deindex_tsv3" found cached.
220429-20:14:55,181 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv4" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv4".
220429-20:14:55,183 nipype.workflow INFO:
[Node] Cached "_deindex_tsv4" - collecting precomputed outputs
220429-20:14:55,183 nipype.workflow INFO:
[Node] "_deindex_tsv4" found cached.
220429-20:14:55,185 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv5" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv5".
220429-20:14:55,186 nipype.workflow INFO:
[Node] Cached "_deindex_tsv5" - collecting precomputed outputs
220429-20:14:55,186 nipype.workflow INFO:
[Node] "_deindex_tsv5" found cached.
220429-20:14:55,188 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv6" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv6".
220429-20:14:55,189 nipype.workflow INFO:
[Node] Cached "_deindex_tsv6" - collecting precomputed outputs
220429-20:14:55,189 nipype.workflow INFO:
[Node] "_deindex_tsv6" found cached.
220429-20:14:55,191 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv7" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv7".
220429-20:14:55,192 nipype.workflow INFO:
[Node] Cached "_deindex_tsv7" - collecting precomputed outputs
220429-20:14:55,192 nipype.workflow INFO:
[Node] "_deindex_tsv7" found cached.
220429-20:14:55,194 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv8" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv8".
220429-20:14:55,195 nipype.workflow INFO:
[Node] Cached "_deindex_tsv8" - collecting precomputed outputs
220429-20:14:55,195 nipype.workflow INFO:
[Node] "_deindex_tsv8" found cached.
220429-20:14:55,197 nipype.workflow INFO:
[Node] Setting-up "_deindex_tsv9" in "/tmp/tmpqslk4cp3/fitlins_wf/deindex_tsv/mapflow/_deindex_tsv9".
220429-20:14:55,198 nipype.workflow INFO:
[Node] Cached "_deindex_tsv9" - collecting precomputed outputs
220429-20:14:55,199 nipype.workflow INFO:
[Node] "_deindex_tsv9" found cached.
220429-20:14:57,242 nipype.workflow INFO:
[Node] Setting-up "_l1_model1" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model1".
220429-20:14:57,300 nipype.workflow INFO:
[Node] Executing "_l1_model1" <fitlins.interfaces.nistats.FirstLevelModel>
tcmalloc: large alloc 1353424896 bytes == 0x2f2aa000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18f8cf5 0x7fedf18f8e08 0x7fedf19b80f4 0x7fedf19bb30c 0x7fedf1b423ac 0x7fedf1b42e10 0x59588e 0x595b69 0x7fedf19c2366 0x4d34e0 0x51286f 0x549576 0x593fce 0x5118f8 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x4bcb19 0x59c019 0x594d69 0x594f8e 0x595509 0x7fedf18f7e54 0x7fedf18fde53 0x7fedf18de24c 0x7fedf18df151 0x7fedf18fdf2a
tcmalloc: large alloc 1353424896 bytes == 0x7fd64000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18f8cf5 0x7fedf18f8e08 0x7fedf19b80f4 0x7fedf19bb30c 0x7fedf1b423ac 0x7fedf1b42e10 0x59588e 0x595b69 0x7fedf19c21e6 0x4d12af 0x5122db 0x549576 0x593fce 0x5118f8 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x4bcb19 0x59c019 0x594d69 0x594f8e 0x595509 0x7fedf18f7e54 0x7fedf18fde53 0x7fedf18de24c 0x7fedf18df151 0x7fedf18fdf2a
tcmalloc: large alloc 2706849792 bytes == 0xd081e000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
tcmalloc: large alloc 1353424896 bytes == 0x2ec36000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18f8cf5 0x7fedf18f8e08 0x7fedf19b80f4 0x7fedf19bb30c 0x7fedf1b423ac 0x7fedf1b42e10 0x59588e 0x595b69 0x7fedf19c2366 0x4d34e0 0x51286f 0x549576 0x593fce 0x5118f8 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x4bcb19 0x59c019 0x594d69 0x594f8e 0x595509 0x7fedf18f7e54 0x7fedf18fde53 0x7fedf18de24c 0x7fedf18df151 0x7fedf18fdf2a
tcmalloc: large alloc 1353424896 bytes == 0x7f6f0000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18f8cf5 0x7fedf18f8e08 0x7fedf19b80f4 0x7fedf19bb30c 0x7fedf1b423ac 0x7fedf1b42e10 0x59588e 0x595b69 0x7fedf19c21e6 0x4d12af 0x5122db 0x549576 0x593fce 0x5118f8 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x4bcb19 0x59c019 0x594d69 0x594f8e 0x595509 0x7fedf18f7e54 0x7fedf18fde53 0x7fedf18de24c 0x7fedf18df151 0x7fedf18fdf2a
tcmalloc: large alloc 2706849792 bytes == 0xd01aa000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
220429-20:16:24,687 nipype.workflow INFO:
[Node] Finished "_l1_model0", elapsed time 91.541693s.
220429-20:16:25,253 nipype.workflow INFO:
[Node] Setting-up "_l1_model2" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model2".
220429-20:16:25,297 nipype.workflow INFO:
[Node] Executing "_l1_model2" <fitlins.interfaces.nistats.FirstLevelModel>
220429-20:16:26,934 nipype.workflow INFO:
[Node] Finished "_l1_model1", elapsed time 89.63268s.
220429-20:16:27,271 nipype.workflow INFO:
[Node] Setting-up "_l1_model3" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model3".
220429-20:16:27,328 nipype.workflow INFO:
[Node] Executing "_l1_model3" <fitlins.interfaces.nistats.FirstLevelModel>
tcmalloc: large alloc 2706849792 bytes == 0x188c30000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
tcmalloc: large alloc 2706849792 bytes == 0x184ca2000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
220429-20:18:14,968 nipype.workflow INFO:
[Node] Finished "_l1_model2", elapsed time 109.670185s.
220429-20:18:15,4 nipype.workflow INFO:
[Node] Finished "_l1_model3", elapsed time 107.674687s.
220429-20:18:15,324 nipype.workflow INFO:
[Node] Setting-up "_l1_model4" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model4".
220429-20:18:15,328 nipype.workflow INFO:
[Node] Setting-up "_l1_model5" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model5".
220429-20:18:15,368 nipype.workflow INFO:
[Node] Executing "_l1_model4" <fitlins.interfaces.nistats.FirstLevelModel>
220429-20:18:15,406 nipype.workflow INFO:
[Node] Executing "_l1_model5" <fitlins.interfaces.nistats.FirstLevelModel>
tcmalloc: large alloc 2706849792 bytes == 0x184ca2000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
tcmalloc: large alloc 2706849792 bytes == 0x188c30000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
220429-20:20:06,152 nipype.workflow INFO:
[Node] Finished "_l1_model5", elapsed time 110.744419s.
220429-20:20:06,994 nipype.workflow INFO:
[Node] Finished "_l1_model4", elapsed time 111.623946s.
220429-20:20:07,445 nipype.workflow INFO:
[Node] Setting-up "_l1_model6" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model6".
220429-20:20:07,452 nipype.workflow INFO:
[Node] Setting-up "_l1_model7" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model7".
220429-20:20:07,496 nipype.workflow INFO:
[Node] Executing "_l1_model6" <fitlins.interfaces.nistats.FirstLevelModel>
220429-20:20:07,500 nipype.workflow INFO:
[Node] Executing "_l1_model7" <fitlins.interfaces.nistats.FirstLevelModel>
tcmalloc: large alloc 2706849792 bytes == 0x1882ec000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
tcmalloc: large alloc 2706849792 bytes == 0x184c94000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
220429-20:22:06,955 nipype.workflow INFO:
[Node] Finished "_l1_model7", elapsed time 119.454213s.
220429-20:22:06,971 nipype.workflow INFO:
[Node] Finished "_l1_model6", elapsed time 119.473432s.
220429-20:22:07,565 nipype.workflow INFO:
[Node] Setting-up "_l1_model8" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model8".
220429-20:22:07,566 nipype.workflow INFO:
[Node] Setting-up "_l1_model9" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model9".
220429-20:22:07,613 nipype.workflow INFO:
[Node] Executing "_l1_model8" <fitlins.interfaces.nistats.FirstLevelModel>
220429-20:22:07,613 nipype.workflow INFO:
[Node] Executing "_l1_model9" <fitlins.interfaces.nistats.FirstLevelModel>
tcmalloc: large alloc 2706849792 bytes == 0x184c64000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
tcmalloc: large alloc 2706849792 bytes == 0x1882ec000 @ 0x7fedf56111e7 0x7fedf18a20ce 0x7fedf18fc726 0x7fedf18fcb09 0x7fedf18fe620 0x7fedf18fed1b 0x7fedf19a041b 0x5936cc 0x548c51 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x549576 0x593fce 0x548ae9 0x5127f1 0x4bc98a 0x532e76 0x595ef6 0x5134a6 0x549576
220429-20:24:01,72 nipype.workflow INFO:
[Node] Finished "_l1_model9", elapsed time 113.457716s.
220429-20:24:03,96 nipype.workflow INFO:
[Node] Finished "_l1_model8", elapsed time 115.479287s.
220429-20:24:26,769 nipype.workflow INFO:
[Node] Setting-up "_l1_model0" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model0".
220429-20:24:26,791 nipype.workflow INFO:
[Node] Cached "_l1_model0" - collecting precomputed outputs
220429-20:24:26,791 nipype.workflow INFO:
[Node] "_l1_model0" found cached.
220429-20:24:26,794 nipype.workflow INFO:
[Node] Setting-up "_l1_model1" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model1".
220429-20:24:26,815 nipype.workflow INFO:
[Node] Cached "_l1_model1" - collecting precomputed outputs
220429-20:24:26,815 nipype.workflow INFO:
[Node] "_l1_model1" found cached.
220429-20:24:26,818 nipype.workflow INFO:
[Node] Setting-up "_l1_model2" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model2".
220429-20:24:26,840 nipype.workflow INFO:
[Node] Cached "_l1_model2" - collecting precomputed outputs
220429-20:24:26,840 nipype.workflow INFO:
[Node] "_l1_model2" found cached.
220429-20:24:26,844 nipype.workflow INFO:
[Node] Setting-up "_l1_model3" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model3".
220429-20:24:26,863 nipype.workflow INFO:
[Node] Cached "_l1_model3" - collecting precomputed outputs
220429-20:24:26,863 nipype.workflow INFO:
[Node] "_l1_model3" found cached.
220429-20:24:26,867 nipype.workflow INFO:
[Node] Setting-up "_l1_model4" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model4".
220429-20:24:26,887 nipype.workflow INFO:
[Node] Cached "_l1_model4" - collecting precomputed outputs
220429-20:24:26,887 nipype.workflow INFO:
[Node] "_l1_model4" found cached.
220429-20:24:26,891 nipype.workflow INFO:
[Node] Setting-up "_l1_model5" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model5".
220429-20:24:26,913 nipype.workflow INFO:
[Node] Cached "_l1_model5" - collecting precomputed outputs
220429-20:24:26,914 nipype.workflow INFO:
[Node] "_l1_model5" found cached.
220429-20:24:26,918 nipype.workflow INFO:
[Node] Setting-up "_l1_model6" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model6".
220429-20:24:26,941 nipype.workflow INFO:
[Node] Cached "_l1_model6" - collecting precomputed outputs
220429-20:24:26,941 nipype.workflow INFO:
[Node] "_l1_model6" found cached.
220429-20:24:26,945 nipype.workflow INFO:
[Node] Setting-up "_l1_model7" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model7".
220429-20:24:26,968 nipype.workflow INFO:
[Node] Cached "_l1_model7" - collecting precomputed outputs
220429-20:24:26,968 nipype.workflow INFO:
[Node] "_l1_model7" found cached.
220429-20:24:26,972 nipype.workflow INFO:
[Node] Setting-up "_l1_model8" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model8".
220429-20:24:26,994 nipype.workflow INFO:
[Node] Cached "_l1_model8" - collecting precomputed outputs
220429-20:24:26,994 nipype.workflow INFO:
[Node] "_l1_model8" found cached.
220429-20:24:26,998 nipype.workflow INFO:
[Node] Setting-up "_l1_model9" in "/tmp/tmpqslk4cp3/fitlins_wf/l1_model/mapflow/_l1_model9".
220429-20:24:27,18 nipype.workflow INFO:
[Node] Cached "_l1_model9" - collecting precomputed outputs
220429-20:24:27,18 nipype.workflow INFO:
[Node] "_l1_model9" found cached.
220429-20:24:49,364 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.collate_run_outputs" in "/tmp/tmpqslk4cp3/fitlins_wf/collate_run_outputs".
220429-20:24:49,373 nipype.workflow INFO:
[Node] Setting-up "_subject_model0" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model0".
220429-20:24:49,389 nipype.workflow INFO:
[Node] Executing "collate_run_outputs" <fitlins.interfaces.utils.CollateWithMetadata>
220429-20:24:49,393 nipype.workflow INFO:
[Node] Finished "collate_run_outputs", elapsed time 0.002556s.
220429-20:24:49,424 nipype.workflow INFO:
[Node] Executing "_subject_model0" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:50,674 nipype.workflow INFO:
[Node] Finished "_subject_model0", elapsed time 1.247832s.
220429-20:24:51,460 nipype.workflow INFO:
[Node] Setting-up "_subject_model1" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model1".
220429-20:24:51,497 nipype.workflow INFO:
[Node] Executing "_subject_model1" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:52,773 nipype.workflow INFO:
[Node] Setting-up "_subject_model2" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model2".
220429-20:24:52,785 nipype.workflow INFO:
[Node] Finished "_subject_model1", elapsed time 1.286648s.
220429-20:24:52,828 nipype.workflow INFO:
[Node] Executing "_subject_model2" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:54,27 nipype.workflow INFO:
[Node] Finished "_subject_model2", elapsed time 1.197272s.
220429-20:24:54,734 nipype.workflow INFO:
[Node] Setting-up "_subject_model3" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model3".
220429-20:24:54,741 nipype.workflow INFO:
[Node] Setting-up "_subject_model4" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model4".
220429-20:24:54,803 nipype.workflow INFO:
[Node] Executing "_subject_model3" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:54,803 nipype.workflow INFO:
[Node] Executing "_subject_model4" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:56,451 nipype.workflow INFO:
[Node] Finished "_subject_model4", elapsed time 1.647018s.
220429-20:24:56,462 nipype.workflow INFO:
[Node] Finished "_subject_model3", elapsed time 1.657688s.
220429-20:24:56,742 nipype.workflow INFO:
[Node] Setting-up "_subject_model5" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model5".
220429-20:24:56,749 nipype.workflow INFO:
[Node] Setting-up "_subject_model6" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model6".
220429-20:24:56,804 nipype.workflow INFO:
[Node] Executing "_subject_model5" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:56,808 nipype.workflow INFO:
[Node] Executing "_subject_model6" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:58,435 nipype.workflow INFO:
[Node] Finished "_subject_model6", elapsed time 1.625546s.
220429-20:24:58,473 nipype.workflow INFO:
[Node] Finished "_subject_model5", elapsed time 1.6678039999999998s.
220429-20:24:58,741 nipype.workflow INFO:
[Node] Setting-up "_subject_model7" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model7".
220429-20:24:58,752 nipype.workflow INFO:
[Node] Setting-up "_subject_model8" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model8".
220429-20:24:58,803 nipype.workflow INFO:
[Node] Executing "_subject_model7" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:24:58,815 nipype.workflow INFO:
[Node] Executing "_subject_model8" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:25:00,502 nipype.workflow INFO:
[Node] Finished "_subject_model7", elapsed time 1.697408s.
220429-20:25:00,539 nipype.workflow INFO:
[Node] Finished "_subject_model8", elapsed time 1.722574s.
220429-20:25:00,741 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts0".
220429-20:25:00,743 nipype.workflow INFO:
[Node] Setting-up "_subject_model9" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model9".
220429-20:25:00,744 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts0" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:00,819 nipype.workflow INFO:
[Node] Executing "_subject_model9" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:25:02,537 nipype.workflow INFO:
[Node] Finished "_subject_model9", elapsed time 1.7168999999999999s.
220429-20:25:02,803 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts0", elapsed time 2.057305s.
220429-20:25:04,752 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts1".
220429-20:25:04,755 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts1" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:05,278 nipype.workflow INFO:
[Node] Setting-up "_subject_model0" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model0".
220429-20:25:05,300 nipype.workflow INFO:
[Node] Cached "_subject_model0" - collecting precomputed outputs
220429-20:25:05,302 nipype.workflow INFO:
[Node] "_subject_model0" found cached.
220429-20:25:05,312 nipype.workflow INFO:
[Node] Setting-up "_subject_model1" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model1".
220429-20:25:05,342 nipype.workflow INFO:
[Node] Cached "_subject_model1" - collecting precomputed outputs
220429-20:25:05,342 nipype.workflow INFO:
[Node] "_subject_model1" found cached.
220429-20:25:05,352 nipype.workflow INFO:
[Node] Setting-up "_subject_model2" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model2".
220429-20:25:05,373 nipype.workflow INFO:
[Node] Cached "_subject_model2" - collecting precomputed outputs
220429-20:25:05,374 nipype.workflow INFO:
[Node] "_subject_model2" found cached.
220429-20:25:05,385 nipype.workflow INFO:
[Node] Setting-up "_subject_model3" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model3".
220429-20:25:05,408 nipype.workflow INFO:
[Node] Cached "_subject_model3" - collecting precomputed outputs
220429-20:25:05,408 nipype.workflow INFO:
[Node] "_subject_model3" found cached.
220429-20:25:05,420 nipype.workflow INFO:
[Node] Setting-up "_subject_model4" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model4".
220429-20:25:05,443 nipype.workflow INFO:
[Node] Cached "_subject_model4" - collecting precomputed outputs
220429-20:25:05,443 nipype.workflow INFO:
[Node] "_subject_model4" found cached.
220429-20:25:05,454 nipype.workflow INFO:
[Node] Setting-up "_subject_model5" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model5".
220429-20:25:05,475 nipype.workflow INFO:
[Node] Cached "_subject_model5" - collecting precomputed outputs
220429-20:25:05,475 nipype.workflow INFO:
[Node] "_subject_model5" found cached.
220429-20:25:05,486 nipype.workflow INFO:
[Node] Setting-up "_subject_model6" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model6".
220429-20:25:05,522 nipype.workflow INFO:
[Node] Cached "_subject_model6" - collecting precomputed outputs
220429-20:25:05,522 nipype.workflow INFO:
[Node] "_subject_model6" found cached.
220429-20:25:05,532 nipype.workflow INFO:
[Node] Setting-up "_subject_model7" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model7".
220429-20:25:05,554 nipype.workflow INFO:
[Node] Cached "_subject_model7" - collecting precomputed outputs
220429-20:25:05,554 nipype.workflow INFO:
[Node] "_subject_model7" found cached.
220429-20:25:05,565 nipype.workflow INFO:
[Node] Setting-up "_subject_model8" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model8".
220429-20:25:05,588 nipype.workflow INFO:
[Node] Cached "_subject_model8" - collecting precomputed outputs
220429-20:25:05,588 nipype.workflow INFO:
[Node] "_subject_model8" found cached.
220429-20:25:05,599 nipype.workflow INFO:
[Node] Setting-up "_subject_model9" in "/tmp/tmpqslk4cp3/fitlins_wf/subject_model/mapflow/_subject_model9".
220429-20:25:05,622 nipype.workflow INFO:
[Node] Cached "_subject_model9" - collecting precomputed outputs
220429-20:25:05,622 nipype.workflow INFO:
[Node] "_subject_model9" found cached.
220429-20:25:06,816 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts1", elapsed time 2.059093s.
220429-20:25:07,59 nipype.workflow INFO:
[Node] Setting-up "_dataset_model0" in "/tmp/tmpqslk4cp3/fitlins_wf/dataset_model/mapflow/_dataset_model0".
220429-20:25:07,93 nipype.workflow INFO:
[Node] Executing "_dataset_model0" <fitlins.interfaces.nistats.SecondLevelModel>
220429-20:25:08,803 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.collate_subject_outputs" in "/tmp/tmpqslk4cp3/fitlins_wf/collate_subject_outputs".
220429-20:25:08,832 nipype.workflow INFO:
[Node] Executing "collate_subject_outputs" <fitlins.interfaces.utils.CollateWithMetadata>
220429-20:25:08,835 nipype.workflow INFO:
[Node] Finished "collate_subject_outputs", elapsed time 0.002034s.
220429-20:25:10,640 nipype.workflow INFO:
[Node] Finished "_dataset_model0", elapsed time 3.546059s.
220429-20:25:11,450 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts2".
220429-20:25:11,451 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts2" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:13,73 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts3".
220429-20:25:13,76 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts3" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:13,111 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts2", elapsed time 1.6586699999999999s.
220429-20:25:14,537 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts3", elapsed time 1.460266s.
220429-20:25:14,756 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.collate_dataset_outputs" in "/tmp/tmpqslk4cp3/fitlins_wf/collate_dataset_outputs".
220429-20:25:14,763 nipype.workflow INFO:
[Node] Executing "collate_dataset_outputs" <fitlins.interfaces.utils.CollateWithMetadata>
220429-20:25:14,763 nipype.workflow INFO:
[Node] Setting-up "_plot_dataset_contrasts0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_dataset_contrasts/mapflow/_plot_dataset_contrasts0".
220429-20:25:14,764 nipype.workflow INFO:
[Node] Finished "collate_dataset_outputs", elapsed time 0.000493s.
220429-20:25:14,765 nipype.workflow INFO:
[Node] Executing "_plot_dataset_contrasts0" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:16,278 nipype.workflow INFO:
[Node] Finished "_plot_dataset_contrasts0", elapsed time 1.511261s.
220429-20:25:18,9 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts4".
220429-20:25:18,10 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts5".
220429-20:25:18,12 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts4" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:18,13 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts5" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:20,320 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts5", elapsed time 2.306176s.
220429-20:25:20,322 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts4", elapsed time 2.309357s.
220429-20:25:20,755 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts6".
220429-20:25:20,757 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts7".
220429-20:25:20,758 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts6" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:20,759 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts7" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:23,59 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts6", elapsed time 2.29996s.
220429-20:25:23,67 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts7", elapsed time 2.306522s.
220429-20:25:24,768 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts8".
220429-20:25:24,769 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts9".
220429-20:25:24,771 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts8" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:24,772 nipype.workflow INFO:
[Node] Executing "_plot_run_contrasts9" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:27,118 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts9", elapsed time 2.344831s.
220429-20:25:27,129 nipype.workflow INFO:
[Node] Finished "_plot_run_contrasts8", elapsed time 2.357118s.
220429-20:25:28,773 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts0".
220429-20:25:28,775 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts0".
220429-20:25:28,776 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts0" - collecting precomputed outputs
220429-20:25:28,776 nipype.workflow INFO:
[Node] "_plot_run_contrasts0" found cached.
220429-20:25:28,778 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts1".
220429-20:25:28,778 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts0" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:28,780 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts1" - collecting precomputed outputs
220429-20:25:28,780 nipype.workflow INFO:
[Node] "_plot_run_contrasts1" found cached.
220429-20:25:28,781 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts2".
220429-20:25:28,783 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts2" - collecting precomputed outputs
220429-20:25:28,783 nipype.workflow INFO:
[Node] "_plot_run_contrasts2" found cached.
220429-20:25:28,784 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts3".
220429-20:25:28,786 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts3" - collecting precomputed outputs
220429-20:25:28,786 nipype.workflow INFO:
[Node] "_plot_run_contrasts3" found cached.
220429-20:25:28,787 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts4".
220429-20:25:28,788 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts4" - collecting precomputed outputs
220429-20:25:28,788 nipype.workflow INFO:
[Node] "_plot_run_contrasts4" found cached.
220429-20:25:28,789 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts5".
220429-20:25:28,791 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts5" - collecting precomputed outputs
220429-20:25:28,791 nipype.workflow INFO:
[Node] "_plot_run_contrasts5" found cached.
220429-20:25:28,792 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts6".
220429-20:25:28,794 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts6" - collecting precomputed outputs
220429-20:25:28,794 nipype.workflow INFO:
[Node] "_plot_run_contrasts6" found cached.
220429-20:25:28,795 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts7".
220429-20:25:28,796 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts7" - collecting precomputed outputs
220429-20:25:28,796 nipype.workflow INFO:
[Node] "_plot_run_contrasts7" found cached.
220429-20:25:28,798 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts8".
220429-20:25:28,799 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts8" - collecting precomputed outputs
220429-20:25:28,799 nipype.workflow INFO:
[Node] "_plot_run_contrasts8" found cached.
220429-20:25:28,800 nipype.workflow INFO:
[Node] Setting-up "_plot_run_contrasts9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_run_contrasts/mapflow/_plot_run_contrasts9".
220429-20:25:28,802 nipype.workflow INFO:
[Node] Cached "_plot_run_contrasts9" - collecting precomputed outputs
220429-20:25:28,802 nipype.workflow INFO:
[Node] "_plot_run_contrasts9" found cached.
220429-20:25:30,259 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts0", elapsed time 1.479549s.
220429-20:25:31,416 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts1".
220429-20:25:31,418 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts1" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:31,418 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts2".
220429-20:25:31,421 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts2" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:33,719 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts1", elapsed time 2.299156s.
220429-20:25:33,729 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts2", elapsed time 2.307207s.
220429-20:25:34,767 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts3".
220429-20:25:34,768 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts4".
220429-20:25:34,769 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts3" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:34,771 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts4" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:37,63 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts3", elapsed time 2.291774s.
220429-20:25:37,82 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts4", elapsed time 2.309764s.
220429-20:25:38,770 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts5".
220429-20:25:38,772 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts6".
220429-20:25:38,773 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts5" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:38,775 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts6" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:41,73 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts5", elapsed time 2.298876s.
220429-20:25:41,93 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts6", elapsed time 2.316509s.
220429-20:25:42,771 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts7".
220429-20:25:42,772 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts8".
220429-20:25:42,774 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts7" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:42,774 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts8" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:45,4 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts7", elapsed time 2.22911s.
220429-20:25:45,57 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts8", elapsed time 2.281283s.
220429-20:25:46,775 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts9".
220429-20:25:46,777 nipype.workflow INFO:
[Node] Executing "_plot_subject_contrasts9" <fitlins.interfaces.visualizations.GlassBrainPlot>
220429-20:25:48,265 nipype.workflow INFO:
[Node] Finished "_plot_subject_contrasts9", elapsed time 1.487576s.
220429-20:25:48,788 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts0" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts0".
220429-20:25:48,789 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts0" - collecting precomputed outputs
220429-20:25:48,790 nipype.workflow INFO:
[Node] "_plot_subject_contrasts0" found cached.
220429-20:25:48,790 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts1" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts1".
220429-20:25:48,791 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts1" - collecting precomputed outputs
220429-20:25:48,791 nipype.workflow INFO:
[Node] "_plot_subject_contrasts1" found cached.
220429-20:25:48,792 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts2" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts2".
220429-20:25:48,793 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts2" - collecting precomputed outputs
220429-20:25:48,793 nipype.workflow INFO:
[Node] "_plot_subject_contrasts2" found cached.
220429-20:25:48,794 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts3" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts3".
220429-20:25:48,794 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts3" - collecting precomputed outputs
220429-20:25:48,795 nipype.workflow INFO:
[Node] "_plot_subject_contrasts3" found cached.
220429-20:25:48,795 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts4" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts4".
220429-20:25:48,796 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts4" - collecting precomputed outputs
220429-20:25:48,796 nipype.workflow INFO:
[Node] "_plot_subject_contrasts4" found cached.
220429-20:25:48,797 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts5" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts5".
220429-20:25:48,798 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts5" - collecting precomputed outputs
220429-20:25:48,798 nipype.workflow INFO:
[Node] "_plot_subject_contrasts5" found cached.
220429-20:25:48,798 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts6" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts6".
220429-20:25:48,800 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts6" - collecting precomputed outputs
220429-20:25:48,800 nipype.workflow INFO:
[Node] "_plot_subject_contrasts6" found cached.
220429-20:25:48,800 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts7" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts7".
220429-20:25:48,801 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts7" - collecting precomputed outputs
220429-20:25:48,801 nipype.workflow INFO:
[Node] "_plot_subject_contrasts7" found cached.
220429-20:25:48,802 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts8" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts8".
220429-20:25:48,803 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts8" - collecting precomputed outputs
220429-20:25:48,803 nipype.workflow INFO:
[Node] "_plot_subject_contrasts8" found cached.
220429-20:25:48,804 nipype.workflow INFO:
[Node] Setting-up "_plot_subject_contrasts9" in "/tmp/tmpqslk4cp3/fitlins_wf/plot_subject_contrasts/mapflow/_plot_subject_contrasts9".
220429-20:25:48,805 nipype.workflow INFO:
[Node] Cached "_plot_subject_contrasts9" - collecting precomputed outputs
220429-20:25:48,805 nipype.workflow INFO:
[Node] "_plot_subject_contrasts9" found cached.
INFO:root:Analysis ID : iarv7
INFO:root:Uploading results to NeuroVault...
Uploading group images
100% 3/3 [00:01<00:00, 2.45it/s]
If you want to run an analysis without Google Drive (i.e. any analysis not using the Budapest dataset), simply remove the --no-get and --dataset-dir arguments, and the fMRI data will be downloaded to the /out directory.
# !neuroscout run --n-cpus=2 $ANALYSIS_ID /out
5) Explore results#
You can explore uploaded results on neuroscout.org by returning to the analysis builder page for your analysis.
Example: https://neuroscout.org/builder/iarv7
You can also view the local analysis outputs, stored in /neuroscout-{ANALYSIS_ID} in this notebook.
!ls /out/neuroscout-${ANALYSIS_ID}/
fitlins options.json sourcedata
The output directory has two sub folders:
fitlins: contains the outputs of the model as executed by FitLinssourcedataincludes all of the inputs required for execution, including Predictor events and the BIDS Stats Model JSON specification. If you did not specificy a cached--dataset-diryou will also find your preprocessed fMRI inputs here.
!ls /out/neuroscout-${ANALYSIS_ID}
fitlins options.json sourcedata
Within the fitlins folder, we have dataset (i.e. group) level result images, HTML analysis reports, as well as subject level results.
For each contrast (in this instance there is only one: speech), we have maps of various statistics available.
Here, we’re going to plot all group level t maps:
from nilearn.plotting import plot_stat_map
from pathlib import Path
outputs = Path(f"/out/neuroscout-{analysis_id}/fitlins/")
statmaps = outputs.glob('level-dataset_*contrast*stat-t*.nii.gz')
for map in statmaps:
plot_stat_map(str(map), title=map.name, threshold=2.58)
/usr/local/lib/python3.7/dist-packages/nilearn/datasets/__init__.py:96: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
"Numpy arrays.", FutureWarning)
/usr/local/lib/python3.7/dist-packages/nilearn/plotting/img_plotting.py:341: FutureWarning: Default resolution of the MNI template will change from 2mm to 1mm in version 0.10.0
anat_img = load_mni152_template()
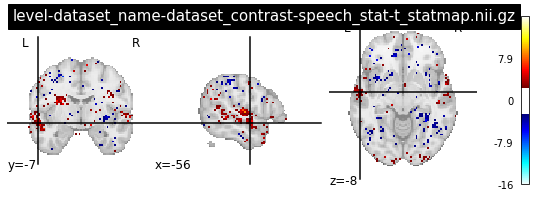
In our example analysis, iarv7, even with only 10 subjects (only first run), we see peak activity in the STS for speech, as expected.

