Ridge regression example#
This notebook implements a cross-valided voxelwise encoding model for a single subject using Regularized Ridge Regression.
The goal is to demonstrate how to obtain Neuroscout data to fit models using custom pipelines. For a comprehensive tutorial, check out the excellent voxelwise modeling tutorials from the Gallant Lab.
Note: By implementing a custom pipeline, your analysis will not be centrally registered on neuroscout.org, and a reproducible record will not be made. For analyses supported the Neuroscout-CLI (e.g. group voxelwise mass univariate GLM models), it is recommended to use the neuroscout.org web interface, or follow the guide for programmatically creating analyses using pyNS.
Citing Neuroscout#
If you publish any results using the Neuroscout data, be sure to cite the Neuroscout paper, and corresponding datasets:
Alejandro de la Vega, Roberta Rocca, Ross W Blair, Christopher J Markiewicz, Jeff Mentch, James D Kent, Peer Herholz, Satrajit S Ghosh, Russell A Poldrack, Tal Yarkoni (2022). *Neuroscout, a unified platform for generalizable and reproducible fMRI research*. eLife 11:e79277. https://doi.org/10.7554/eLife.79277
Visconti di Oleggio Castello, M., Chauhan, V., Jiahui, G., & Gobbini, M. I. An fMRI dataset in response to “The Grand Budapest Hotel”, a socially-rich, naturalistic movie. Sci Data 7, 383 (2020). https://doi.org/10.1038/s41597-020-00735-4
If running this notebook on Google Colab, be sure to switch the runtime to GPU to expedite model fitting (Runtime -> Change runtime type)
#@title 1) Run once to install dependencies on Colab (pyNS, Datalad, and Machine Learning libraries) (~1 minute)
%%capture --no-display --no-stderr
from IPython.display import display
display("Installing NeuroDebian...")
## Set up DataLad
!wget -O- http://neuro.debian.net/lists/focal.us-ca.libre | sudo tee /etc/apt/sources.list.d/neurodebian.sources.list && sudo apt-key adv --recv-keys --keyserver hkps://keyserver.ubuntu.com 0xA5D32F012649A5A9 && sudo apt-get update
display("Installing DataLad...")
!sudo apt-get install datalad -y
%pip install -U datalad
!git config --global user.email "you@example.com" && git config --global user.name "Your Name"
display("Installing PyNS...")
%pip install pyns
%pip install git+https://github.com/bids-standard/pybids.git#egg=pybids
display("Installing analysis dependencies (nilearn, and himalaya)...")
%pip install nilearn himalaya
display("Done.")
UsageError: Line magic function `%%capture` not found.
Fetching Neuroscout Data#
We can easily retrieve data from Neuroscout using pyNS — the Python Neuroscout API client.
Be sure to refer to the official pyNS documentation for further usage information, with particular focus on the section on fetching predictors and images.
What datasets are available?#
If you’re not sure what is available, you can browse Neuroscout’s datasets and predictors online.
Here, we were going to focus on the ‘Budapest’ dataset, choosing the first subject (sid000005) as an example.
# Find first subject for Budapest
from pyns import Neuroscout
api = Neuroscout()
dataset_name = 'Budapest'
# First run for Budapest dataset
first_run = api.runs.get(dataset_name=dataset_name)[0]
subject = first_run['subject'] # Save subject name
print(subject)
sid000005
Fetching predictors#
We will fetch two sets of predictors: Mel spetrogram, and Mel Frequency Cepstral Coefficient (MFCC). Both of these features are extracted from the auditory track of the movie stimulus (‘The Grand Budapest Hotel’). Later in the tutorial we’ll fit an encoding model to each set of features separately.
First, we define the names of the predictors we will fetch.
To learn more about basic Neuroscout API querying, see this guide.
mfccs = [f'mfcc_{i}' for i in range(20)]
mel = [f'mel_{i}' for i in range(64)]
all_vars = mfccs + mel
Next, we can use the high-level utility fetch_predictors to retrieve these predictors for the target subject, rescaled to unit variance, and resampled to the imaging data’s Repetition Time (TR).
Since downloading and resampling can take a minute, for this example we’ll only use the first three (out of 8) runs.
from pyns.fetch_utils import fetch_predictors
predictors = fetch_predictors(all_vars, dataset_name='Budapest', subject='sid000005', run=[1, 2, 3], rescale=True, resample=True)
This results in a DataFrame with predictors, plus metadata such as file entities (e.g. subjects, runs)
Fetch fMRI data and loading images#
To retrieve Neuroscout data, we use datalad to fetch the preprocessed images remote.
pyNS includes a helper function to facilitate installing and fetching the dataset using datalad: fetch_images.
Provide the name of the dataset (Budapest), plus a directory where your datasets are stored, plus (optionally) filters to restrict which files are downloaded (e.g. subject)
from pyns.fetch_utils import fetch_images
# Note: This command can hang after fetching images.
# If so, stop this cell and re-run, and it will immediately re-execute
preproc_dir, img_objs = fetch_images('Budapest', '/tmp/', subject=subject, run=[1, 2, 3], fetch_brain_mask=True)
/home/alejandro/miniconda3/envs/nsencoding/lib/python3.10/site-packages/bids/layout/validation.py:48: UserWarning: The ability to pass arguments to BIDSLayout that control indexing is likely to be removed in future; possibly as early as PyBIDS 0.14. This includes the `config_filename`, `ignore`, `force_index`, and `index_metadata` arguments. The recommended usage pattern is to initialize a new BIDSLayoutIndexer with these arguments, and pass it to the BIDSLayout via the `indexer` argument.
warnings.warn("The ability to pass arguments to BIDSLayout that control "
/home/alejandro/miniconda3/envs/nsencoding/lib/python3.10/site-packages/bids/layout/validation.py:122: UserWarning: The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
warnings.warn("The PipelineDescription field was superseded "
action summary:
get (notneeded: 6)
The fetch_images function returns images as pybids BIDSImageFile objects, which include metadata such as entities as part of the object
img_objs[2].get_entities()
{'datatype': 'func',
'desc': 'preproc',
'extension': '.nii.gz',
'run': 3,
'space': 'MNI152NLin2009cAsym',
'subject': 'sid000005',
'suffix': 'bold',
'task': 'movie'}
Using the entities, we can separate the list of images into two lists: preprocessed functional images and brain masks:
all_funcs = [f for f in img_objs if f.entities['suffix'] == 'bold']
all_masks = [f for f in img_objs if f.entities['suffix'] == 'mask']
Image preprocessing#
The following computes a joint mask for all runs. Alternatively, we could also provide a apriori ROI mask here.
import nibabel as nib
import numpy as np
def _compute_mask_intersection(masks):
""" Compute joint masks (i.e. where all masks have 1s)"""
_masks = [nib.load(m) for m in masks]
_data = np.stack(n.get_fdata() for n in _masks)
intersection = _data.min(axis=0)
mask = nib.Nifti1Image(
intersection, affine = _masks[0].affine, header=_masks[0].header)
return mask
inter_mask = _compute_mask_intersection(all_masks)
/home/alejandro/miniconda3/envs/nsencoding/lib/python3.10/site-packages/IPython/core/interactiveshell.py:3442: FutureWarning: arrays to stack must be passed as a "sequence" type such as list or tuple. Support for non-sequence iterables such as generators is deprecated as of NumPy 1.16 and will raise an error in the future.
exec(code_obj, self.user_global_ns, self.user_ns)
Now we can apply the corresponding brain mask to each run using NiftiMasker from nilearn, and stack them into a single array for subsequent analysis:
import pandas as pd
import numpy as np
from nilearn.maskers import NiftiMasker
def _mask_and_stack_images(image_objects, mask):
""" Stack images into single array, and collect metadata entities into DataFrame """
masker = NiftiMasker(mask_img=mask)
arrays = []
entities = []
image_objects = sorted(image_objects, key=lambda x: x.entities['run'])
for img in image_objects:
run_y = masker.fit_transform(img)
arrays.append(run_y)
entities += [dict(img.entities)] * run_y.shape[0]
entities = pd.DataFrame(entities)
return np.vstack(arrays), entities, masker
y, img_entities, masker = _mask_and_stack_images(all_funcs, inter_mask)
The stacked runs have shape: (n_volumes, n_voxels)
y.shape
(1631, 124614)
We also keep the entities associated with each volume in a dataframe:
img_entities
| datatype | desc | extension | run | space | subject | suffix | task | |
|---|---|---|---|---|---|---|---|---|
| 0 | func | preproc | .nii.gz | 1 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 1 | func | preproc | .nii.gz | 1 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 2 | func | preproc | .nii.gz | 1 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 3 | func | preproc | .nii.gz | 1 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 4 | func | preproc | .nii.gz | 1 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1626 | func | preproc | .nii.gz | 3 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 1627 | func | preproc | .nii.gz | 3 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 1628 | func | preproc | .nii.gz | 3 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 1629 | func | preproc | .nii.gz | 3 | MNI152NLin2009cAsym | sid000005 | bold | movie |
| 1630 | func | preproc | .nii.gz | 3 | MNI152NLin2009cAsym | sid000005 | bold | movie |
1631 rows × 8 columns
Fitting ridge regression models using Mel spectrogram features#
First, we’ll fit a basic voxelwise ridge regression model to a subset of voxels (for demonstration purposes).
We’ll define two cross-validators: an outer and an inner CV. The outer cross-validator will loop will be used to estimate the performance of the model on unseen data, and the inner cross-validator will be used to select the alpha hyperparameter for ridge regression, within each training fold of the outer cross-validator.
In both cases, we’ll use the leave-one-run-out strategy, testing the model on an unseen run. Since there are multiple observations per run, we use therun column of X_entities to group observations, and ensure they appear together within each fold.
Finally, we also define a scoring function, in this case, correlation_score.
Note that we are using the himalaya library’s KernelRidgeCV estimator, as it is optimized for multi-target (i.e. voxel) estimation & hyperparameter optimization with an API inspired by scikit-learn.
from himalaya.ridge import RidgeCV
from sklearn.model_selection import GroupKFold
from himalaya.scoring import correlation_score
from himalaya.backend import set_backend
backend = set_backend("torch_cuda", on_error="warn")
groups = predictors['run'].tolist()
# Set up estimator and CV objects
estimator = RidgeCV()
n_runs = len(set(groups))
cv = GroupKFold(n_splits=n_runs)
inner_cv = GroupKFold(n_splits=n_runs - 1)
/home/alejandro/miniconda3/envs/nsencoding/lib/python3.10/site-packages/himalaya/backend/_utils.py:56: UserWarning: Setting backend to torch_cuda failed: PyTorch with CUDA is not available..Falling back to numpy backend.
warnings.warn(f"Setting backend to {backend} failed: {str(error)}."
The following is a generic pipeline for applying the estimator to the data:
from collections import defaultdict
def _model_cv(estimator, cv, X_vars, y, groups=None,
scoring=correlation_score, inner_cv=None):
""" Cross-validate a model on a set of variables.
Parameters
----------
estimator : Scikit-learn like estimator
Estimator to use for fitting the model. Must have a fit and predict
method.
cv : cross-validation generator or an iterable
Outer cross-validation generator to use for cross-validation.
X_vars : pandas.DataFrame
Dataframe containing the variables to use for prediction.
y : array-like, shape (n_samples, n_targets)
Target values.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
scoring : callable, optional (default: correlation_score)
Scoring function to use for evaluating the model.
inner_cv : cross-validation generator or an iterable, optional
Inner cross-validation generator to use for cross-validation.
"""
# Container for results
results = defaultdict(list)
# Extract data from dataframe
X = X_vars.values
# Extract number of samples for convenience
n_samples = y.shape[0]
# Loop through outer cross-validation folds
for train, test in cv.split(np.arange(n_samples), groups=groups):
# Get training model for list of model bands
X_train = [x[train] for x in X] if type(X) == list else X[train]
X_test = [x[test] for x in X] if type(X) == list else X[test]
# Create inner cross-validation loop if specified
if inner_cv:
# Split inner cross-validation with groups if supplied
inner_groups = np.array(groups)[train] if groups else groups
inner_splits = inner_cv.split(np.arange(n_samples)[train],
groups=inner_groups)
# Update estimator with inner cross-validator
estimator.set_params(cv=inner_splits)
# Fit the regression model on training data
estimator.fit(X_train, y[train])
# Compute predictions
test_prediction = estimator.predict(X_test)
# Test scores
test_score = scoring(y[test], test_prediction)
# If output is not an array, assume its a torch Tensor and convert to array
if not isinstance(test_prediction, np.ndarray):
test_prediction = test_prediction.cpu().numpy()
test_score = test_score.cpu().numpy()
# Populate results dictionary
results['test_predictions'].append(test_prediction)
results['test_scores'].append(test_score)
# Combine into single aray
results['test_scores'] = np.stack(results['test_scores'])
return results
Mel-spectrogram features only#
First, we’ll fit this model using only the Mel spectrogram features:
X = predictors[mel] # Take a subset of X for this model
results = _model_cv(estimator, cv, X, y, inner_cv=inner_cv, groups=groups)
The results dictionary contains:
print(results.keys())
dict_keys(['test_predictions', 'test_scores'])
The test_scores and coefficients are of shape: (n_folds, n_voxels).
results['test_scores'].shape
(4, 124614)
Then, we average across folds to get the mean score across for this subject:
mean_scores = results['test_scores'].mean(axis=0)
For interpretation only, we’ll only look at positive scores:
mean_scores[mean_scores < 0] = 0
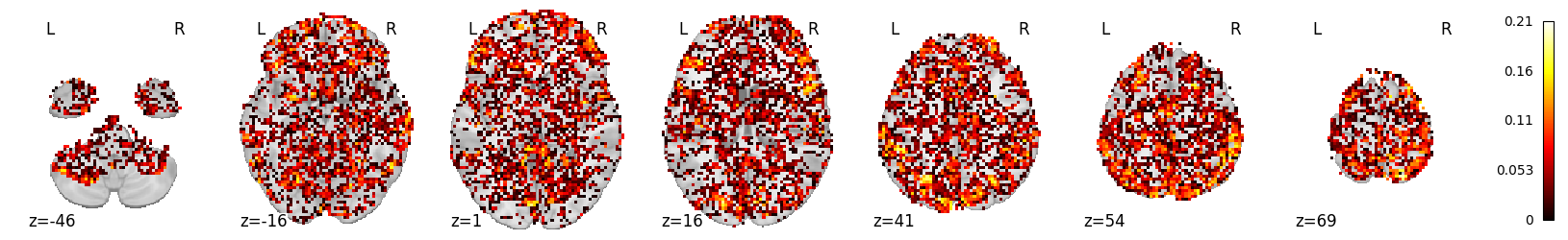
mean_scores.max()
0.21056749957050686
mean_scores.mean()
0.027309387725607995
We can inverse transform the score map, to a 3D volume, and vizualize it:
from nilearn.plotting import plot_stat_map
plot_stat_map(masker.inverse_transform(mean_scores), display_mode='z')
<nilearn.plotting.displays._slicers.ZSlicer at 0x7f44051f3b50>
# Clean up (for memory consumption)
del results['test_scores']
del results
Ridge regression using MFCC features#
We can now run a similar model, but using MFCC featues, instead of the Mel spectrogram
X = predictors[mfccs]
results_mfcc = _model_cv(estimator, cv, X, y, inner_cv=inner_cv, groups=groups)
mean_scores_mfcc = results_mfcc['test_scores'].mean(axis=0)
mean_scores_mfcc[mean_scores_mfcc < 0] = 0
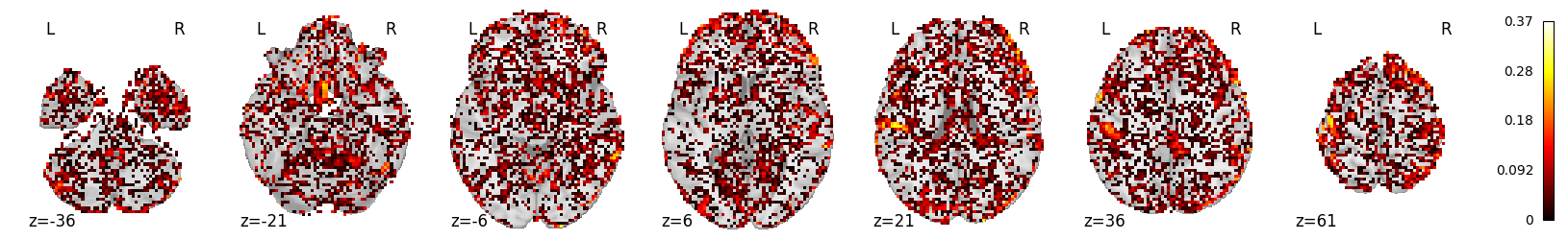
From looking at whole-brain scores, the MFCC mdoel may outperform the Mel spectrogram model:
mean_scores_mfcc.max()
0.36887338343916876
mean_scores_mfcc.mean()
0.02817772388516969
from nilearn.plotting import plot_stat_map, plot_glass_brain
plot_stat_map(masker.inverse_transform(mean_scores_mfcc), display_mode='z')
<nilearn.plotting.displays._slicers.ZSlicer at 0x7f4407c4e020>
# Clean up (for memory consumption)
del results_mfcc['test_scores']
del results_mfcc
Finite impulse response model#
Due to the hemodynamic lag, the model may perform better if the predictors were delayed relative to the stimulus onset.
We can do so using a Finite Impulse Response (FIR) model.
Using fetch_predictors, we can ask for the predictors to be returned as a BIDSVariableCollection, which enables us to apply any of the transformations implemented in pybids.
Alternatively, you could apply any arbitrary transformations, by requesting a pandas DataFrame.
collection = fetch_predictors(all_vars, dataset_name='Budapest', subject='sid000005',
rescale=True, resample=True, return_type='collection')
Next, we use Convolve to apply a fir model, and convert to a pandas df
from bids.modeling.transformations import Convolve
def _convolve_df(collection, predictor_names):
Convolve(collection, predictor_names, model='fir', fir_delays=[1, 2, 3, 4])
# To df, and sort rows by keys
collection_df = collection.to_df().sort_values(['subject', 'run', 'onset'])
# Reorder columns
sort_columns = ['onset', 'duration'] + predictor_names
sort_columns += collection_df.columns.difference(sort_columns).tolist()
collection_df = collection_df[sort_columns]
return collection_df
collection_df = _convolve_df(collection, all_vars)
collection_df.shape
(3052, 425)
The FIR convolution results in 5x amount of predictors, including the delayed versions of the predictors.
We can now fit a model with all the Mel spectrogram features, to see if it improves prediction:
X = collection_df.iloc[:, collection_df.columns.str.startswith('mel')]
results_mel_fir = _model_cv(estimator, cv, X, y, inner_cv=inner_cv, groups=groups)
mean_scores_mel_fir = results_mel_fir['test_scores'].mean(axis=0)
mean_scores_mel_fir[mean_scores_mel_fir < 0] = 0
The max score increases, while the mean score remains similar (although marginally higher):
mean_scores_mel_fir.max()
0.28626928535573065
mean_scores_mel_fir.mean()
0.026594240316020888
plot_stat_map(masker.inverse_transform(mean_scores_mel_fir), display_mode='z')
<nilearn.plotting.displays._slicers.ZSlicer at 0x7f4407769570>
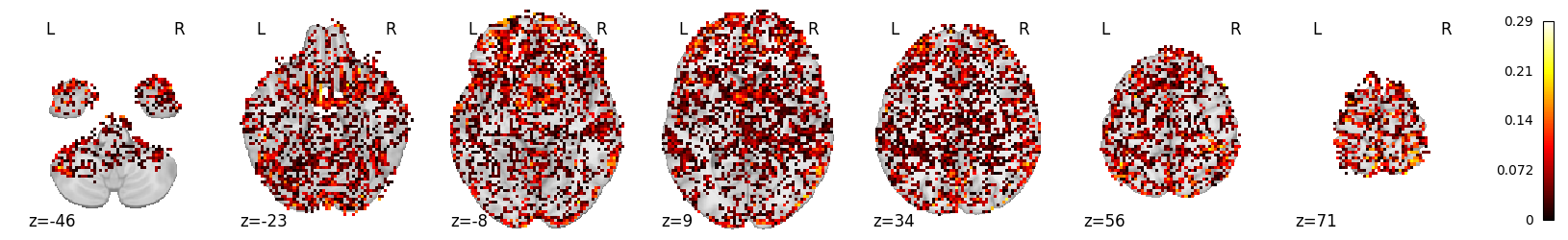
Where to go from here?#
The goal of this tutorial is to familiarize you with how to access Neuroscout’s data to fit custom models.
This tutorial does not exhaustively cover all possible models that can be fit using these data, but at this point you should have the tools necessary to access the necessary data from NeuroScout’s API.
By being able to fetch both predictor and brain imaging time courses using a simple, uniform API, it should be easy to extend these examples to a wide variety of machine learning workflows.
Citing Neuroscout#
Please ensure to cite Neuroscout if publishing any work using these data, and be aware that there are ongoing efforts to standardize the most common variations of voxel-wise encoding models, in order to enable users to fully specify and register their analysis in Neuroscout (like you currently can for summary-statistics multi-level GLM models).

